
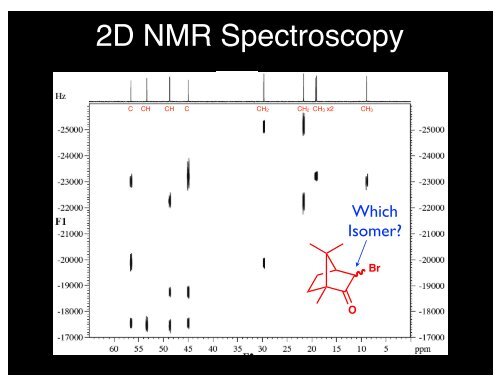
In a first step, mono- and bidimensional SSNMR spectra, i.e., 1H Magic-Angle Spinning (MAS), 13C and 15N Cross Polarisation Magic-Angle Spinning (CPMAS), 1H Double Quantum (DQ) MAS, 1H- 13C HETeronuclear CORrelation (HETCOR), were used to determine the correct molecular structure (i.e., zwitterionic or not) and the local molecular arrangement at the end, the RMSEs between experimental and computed 1H and 13C chemical shifts allowed the selection of the correct predicted structure for each system. In this paper, we present the successful application of CSP-NMRX to determine the crystal structure of three structural isomers of pyridine dicarboxylic acid, namely quinolinic, dipicolinic and dinicotinic acids, which can be in a zwitterionic form, or not, in the solid state. Furthermore, it is well known that the coupling of CSP with solid-state NMR (SSNMR) greatly enhances the performance and the accuracy of the predictive method, leading to the so-called CSP-NMR crystallography (CSP-NMRX).

When it comes to crystal structure determination, computational approaches such as Crystal Structure Prediction (CSP) have gained more and more attention since they offer some insight on how atoms and molecules are packed in the solid state, starting from only very basic information without diffraction data.


 0 kommentar(er)
0 kommentar(er)
